About Our Project
Honours Project
Modelling the human cell cycle: developing a dynamically generated user interface
The project developed an easy-to-use interface to an existing biological pathway model to allow medical biologists to use that model. The previous interface was entirely text based. During requirements analysis it was identified that a static interface to a single model would not be appropriate. As new information became available the model changed both its shape and parameters. Thus a user interface that is generated dynamically based on the latest model was developed. The interface allows a user to create, load, edit, save and execute biological pathway models with no computer programming. Due to the multidisciplinary nature of the project some background information is required to explain the work undertaken.
Background information
Biological Pathway models attempt to describe cellular processes and regulatory networks by reproducing observed cell behaviour, such as protein and other chemical concentrations, gross behaviour of cells such as mass at division or apoptosis (cell death) during the cell cycle. It is believed that by applying the models to specific cell processes that experimental results for these processes can be predicted.
At current rates, about one in four of us will die from cancer, which is responsible for 127,000 deaths every year in the UK. As cancer cells are caused by the malfunction of the cell cycle processes, it is these processes and the other more complex systems that control these processes, which the models attempt to reproduce. It is hoped that by modelling the cell cycle regulatory systems successfully that the effects of cancer treatment methods can be tested on the models and more successful treatment regimes can be found for specific patients and cancer types. For example if a biopsy taken from a lung cancer patient shows that the cancer cells lack certain protein complexes which could prevent the cells dividing then a model could be used to determine which combination of drugs would have the greatest effect on stopping or killing the cancer cells. This could greatly reduce the drug development process since a potential solution can be identified before undertaking the usual testing process, and so reducing the possibility of administering harmful drug combinations.
The Current System
Biological pathway models obviously have great potential for the medical community by allowing the development of drug treatment regimes for specific types of cancer cell and for specific patients. These models consider the cell in terms of interacting parts, connected by flows of chemicals.
A PhD project at the University of Abertay’s SIMBIOS environmental research centre created a model of the human cell cycle the represents many aspects of cell behaviour. The model is composed of the cell cycle representation, including phase (system state) variables that represent the values of key proteins, a differential equation set that link different components and hundreds of parameters for that set, and ancillary tools including a genetic algorithm search process and the integration solver needed for the equation solutions. The model components can be changed to reflect the type of cell or process being investigated. The current system uses the Matlab software’s scripting language to create and edit the models and their components. While still in the development stage the model successfully represents the required cell processes.
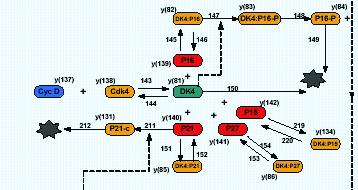
The cell processes are modelled using a set of non-linear differential equations (dx/dt = f(x,t,c)) , where x is a set of phase variables, t is a given time period and c is a set of parameters. A generic form of the model uses over 85 differential equations and a parameter set of about 300 variables. A more specific form of the model to test the effects of drug treatments on a specific cell type would use a much larger parameter set and many more equations. These equations are passed through a solver coded with the Matlab software. The solver combines a numerical integrator, a genetic algorithm and a set of fitness functions, which attempts to find the best parameter set c, for a given value of x.
Rational for a Graphical User Interface
There was however a large problem with the design of the models. The models were created and edited by hand in the form of m.files, Matlab’s scripting files. Each model’s m-file could potentially contain hundreds (if not thousands) of lines of code specifying everything from the model’s name to the form of the genetic algorithm used.
The problem is that for the models created to be of any use in medical research and the development of cancer treatments it must be possible for a person from a biological or medical background to use it. Currently this is not the case, as the user would have to have extensive knowledge of the Matlab language and its operation to even attempt to change a single variable. The user would also have to have knowledge of the solver and fitness function used to enable their application to a different model. With out some kind of simpler interface someone other than the designer of the algorithms, solver and fitness functions used could not use the models, in any practical way.
Key Functions of the Application
Due to the complexity and stability issues of the current model it would not be possible to simply recode the model in another language to produce a model with an interface. To overcome this problem the GUI was designed to run as a standalone application, which can interface with the Matlab software. The operation of the GUI in this way allows the models internal code and operation to be completely abstracted away from the user. The Java Programming language was selected because of its flexibility in the creation of graphical interfaces and also its portability across various platforms.
The primary function of the application is that it allows a user unfamiliar with the mathematical code and the Matlab language and operation to create, load, edit, save and execute a biological pathway model. The models components are displayed in a hierarchal tree structure, which allows easy identification of the separate types of components that make up the models functionality. Each model component is editable by selecting its node in the tree. The application allows the user to add and remove model components at will through the use of a simple right click context menu. When a component is selected in the tree a unique panel will be displayed for that type of component. Phase variable and equation expressions are entered using a drag & drop equation editor, preventing the user entering an expression containing other components which do not exist. The application also allows the user to enter comments about each model component. The comments may simply contain text describing that component, but can also contain direct hyperlinks to research papers, other documents and WebPages. Activated links are passed to the systems default browser, this means that the application doesn’t need to contain any extra file handling code.
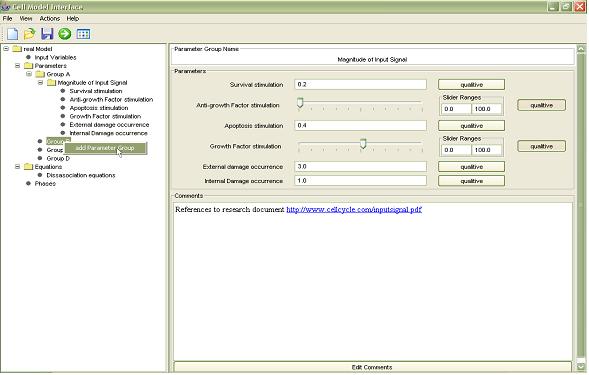
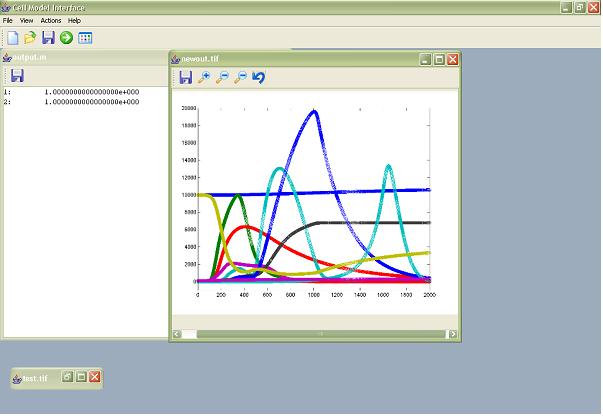
The user is able to save the created model’s m-file, i.e. the interface can create and/ or modify an existing Matlab script through interactions, for later use or execute it directly on the Matlab platform. The Matlab execution of the m-file is all done in the background so that the user is never presented with program code and the state of the Matlab environment such as run-time information. While the model is running the user will be able to view the results, i.e. the system-level phase variables, being depicted in real-time via a constantly updated image. When the model run has finished the user has the option of resizing, adding notes or axis labels to the image and allows the user to save the produced image as a tiff file, again all through the use of a simple context menu. The application will also display the numerical results to the user, which can also be saved if desired. Should any errors occur in the running of the script the user will be informed by the application and Matlab will automatically be shut down.
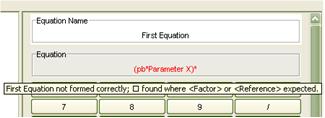
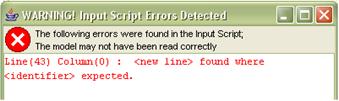
Through the use of recursive descent compiler technology, the application provides extensive m-file error validation. When a script is saved, executed or opened, the user will be informed of any errors founding the script. However they will not be prevented from saving or executing as the model may be a work in progress or as it is a research tool certain effects may be desirable. The application will attempt to read in as much of an incorrect m-file as possible to enable the user to retain any model components that were correctly specified. Should any errors be found the user will be presented with a list of errors detailing the line and column in the m-file on which the error was detected or the equation or phase variable in which the error occurred. The application also includes error highlighting for equation and phase variable expressions, the text of the expression will turn red and the error detected will be displayed as a tool tip.
Summary
The underlying computational model of the cell cycle will continue to change. However, the approach used – that of a set of differential equations, parameters to those equations and system-level phase variables will remain constant. Importantly, the developed software is generic and able to reflect those changes in the interface on screen. As a result, the interface and the code behind it will continue to be used for the foreseeable future with newer or more advanced models.
